Automatic Mushroom Farm
I have gone about this project before and want to set it up again, hoping to use something like this. But using water to retain the optimal humidity instead so it would not need a separate humidifier.
Bioinformatician & Researcher
Exploring the intersection of academia, biostatistics, privacy, and outdoor pursuits

I am Mike Loomans, an industrious bioinformatician, aspiring to contribute significantly to my field and beyond. My interests span the realms of academia, biostatistics, privacy, and outdoor pursuits.
While my numerous side projects may fade into obscurity once completed, this page serves as a testament to my commitment to maintaining transparency and organization throughout the entire project lifecycle—from inception to final production.
Recognizing that some of my ideas may be considered ambitious, I find it essential to document them, acknowledging the potential utility that may arise as my skills and technical capabilities continue to evolve.

I have gone about this project before and want to set it up again, hoping to use something like this. But using water to retain the optimal humidity instead so it would not need a separate humidifier.
During the 2025 BioSB conference I met a ton of interesting people and listened to a great number of cool talks. While there I presented my package on the creation of bioprospecting models as pip package called migenpro in the form of a demo.
Genome-wide association studies are on the rise, thanks to the continuous development of novel sequencing techniques. This trend underscores the demand for appropriate tools and analyses. One promising avenue within GWAS involves phenotype prediction, wherein machine learning models can anticipate phenotypes and traits, including substances like 1,3 biopropanol.
The global scale of pig production has reached massive proportions and continues to grow, driven by increasing incomes in the developing world. However, intensive livestock farming and the conditions needed for profitability contribute to the spread of infectious diseases, such as Post-Weaning Diarrhea (PWD), particularly in industrial pig farming.
My role in this project focuses on the microbiome analysis of the piglet gut post weaning. To accomplish this goal I envision a graph database capable of showing the intertwined nature of all genomes within the pig gut.
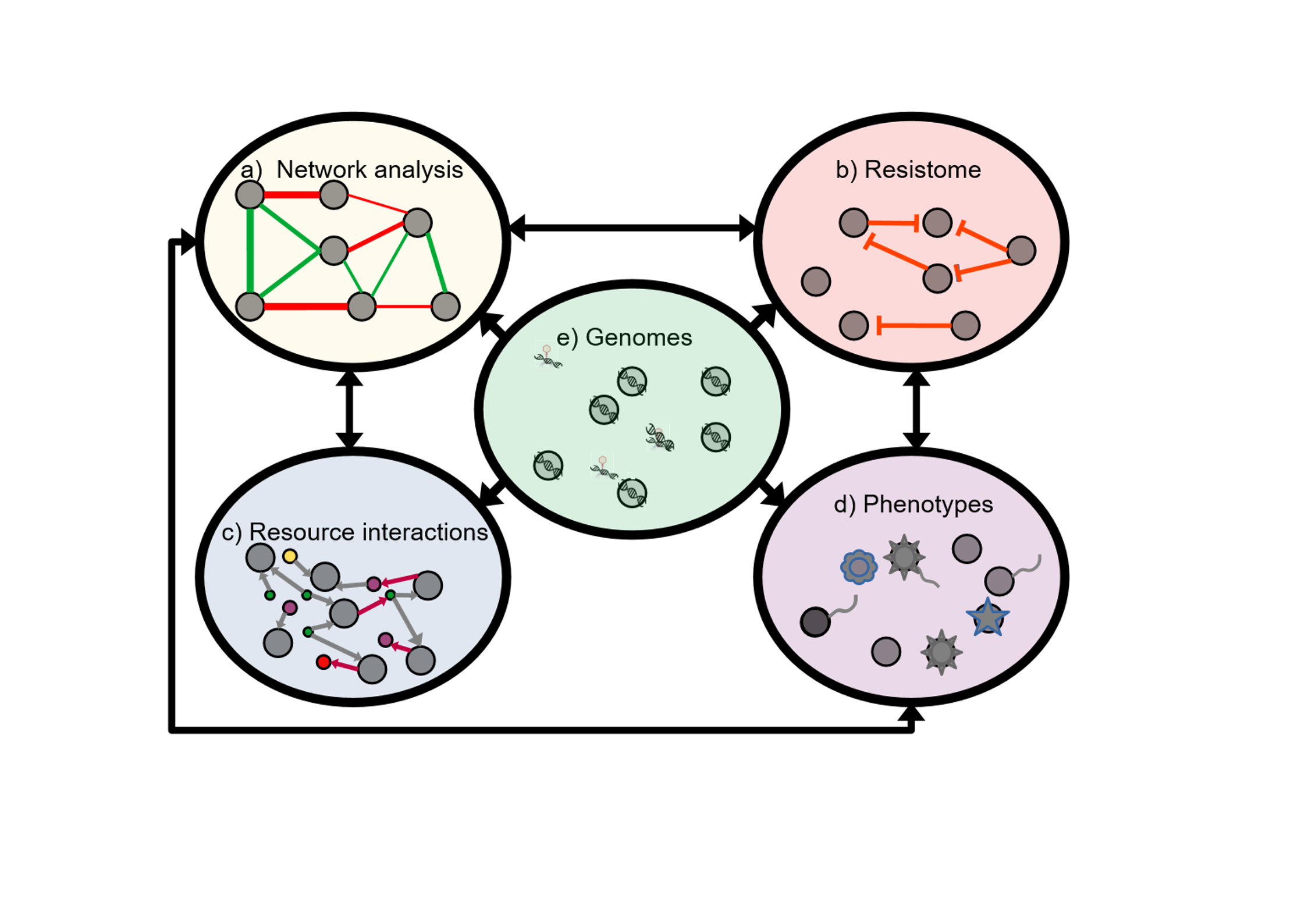
Development of a viromics pipeline geared towards the analysis of the SHIME system
x.y@wur.nl
x = mike
y = loomans