Speleology
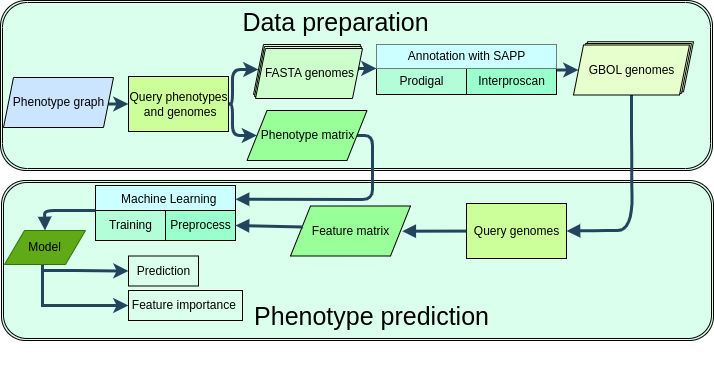
The increasing availability of microbial genomic data and recent development of machine learning and AI methods create a unique opportunity to establish associations between genetic information and phenotypes. Here we present a computational framework for Microbial Genome Prospecting (Migenpro), that combines phenotype and genomic linked data.
Migenpro serves as a framework for the generation of machine learning models that predict microbial traits from genome sequences. Microbial whole genome sequences have been consistently annotated and genomic features were stored in a semantic framework using the Genome Biology Ontology Language (GBOL). Phenotype data corresponding to the available genome sequences was retrieved from BacDive and the associations between phenotype and genotype were used to train machine learning models.
Our approach successfully predicts traits such as motility, Gram stain, optimal growth temperature range, and sporulation capabilities. To ensure robustness, five-fold cross-validation was implemented and demonstrated consistent model performance across iterations and did not indicate overfitting.
The framework's effectiveness was further validated through comparison with previously published models, showing comparable accuracy, with modest variations attributed to differences in datasets rather than methodology. The classification models can be further explored using feature importance characterization to identify biologically relevant genomic features. Migenpro provides an interoperable framework to predict phenotypes from genomic data.